

Molecular Biology Division is engaged in obtaining the mechanistic insights into mechanisms important for key cellular functions, and the ability to tolerate various abiotic and biotic stresses.
Major objectives:
1) Delineation of the role of proteins forming part of DNA repair interactome and secondary structures in DNA in radiotolerance in bacteria.
2) Involvement of Serine/Threonine (Ser/Thr) kinases, nucleotide kinases and extracellular DNA in bacterial cell physiology and stress tolerance.
3) Understanding interaction of metals with microbes, and exploring utility of important mechanisms in bioremediation.
4) Role of hypothetical intrinsically disordered proteins in eukaryotic cell functions and bacterial stress physiology.
5) Understanding role of epigenetic regulatory mechanisms crucial in cancer.
6) Deciphering molecular basis of structural-functional diversity of key epigenetic regulators in plants.
7) Biomarker studies for developing robust tool for cancer detection using machine learning (ML).
Alternative splicing (AS), a post-transcriptional regulatory mechanism, contributes significantly in enhancement of transcriptome/proteome diversity. AS via generating multiple transcripts by various types of AS events that may also encode alternative protein isoforms with novel characteristics. Copper-Zinc Superoxide dismutases (CSDs) in rice display extensive alternative splicing events leading to varying levels of constitutive and alternative transcripts under environmental stress conditions, in different tissues. Furthermore, alternative splicing also targets the coding regions of the SOD genes, affecting key sites/regions crucial for structure and function of the alternatively spliced protein isoforms.
The Mn-catalase, KatB, plays a vital role in adaptation to salinity and oxidative stress in the cyanobacterium Anabaena. Biochemically, KatB is a thermostable, robust Mn-catalase that functions at alkaline pH. The KatB crystal structure, the first one from a photosynthetic organism, showed its active site to be distinct from other Mn-catalases. The N-terminal region of this enzyme was shown to play a crucial role in subunit interaction and maintaining the proper active site geometry. Very recently, KatB was purified from the salt-stressed wild-type Anabaena, showcasing a non-recombinant route of obtaining this catalase.
Protein diversity in a mammalian cell is generated from various mechanisms, alternative splicing of mRNA is one of them. BRD4 is an epigenetic regulator and a transcription factor, belongs to the BET (Bromodomain and extra terminal domain) family. BRD4 protein has been implicated in inducing oncogenic phenotypes in many cancers. BRD4 is of particular interest in cancer therapeutics as it is a potential druggable target for cancer treatment. Number of BET inhibitors, which act by blocking Bromodomain binding to chromatin, are being tested for their anti-cancer properties in FDA approved Phase I/II clinical trials. We have found that BRD4 deficiency alters patterns of transcript splicing both in normal thymocytes and in cancer cells by interacting with components of the spliceosome complex.
For studying macromolecular interactions and particle size estimations
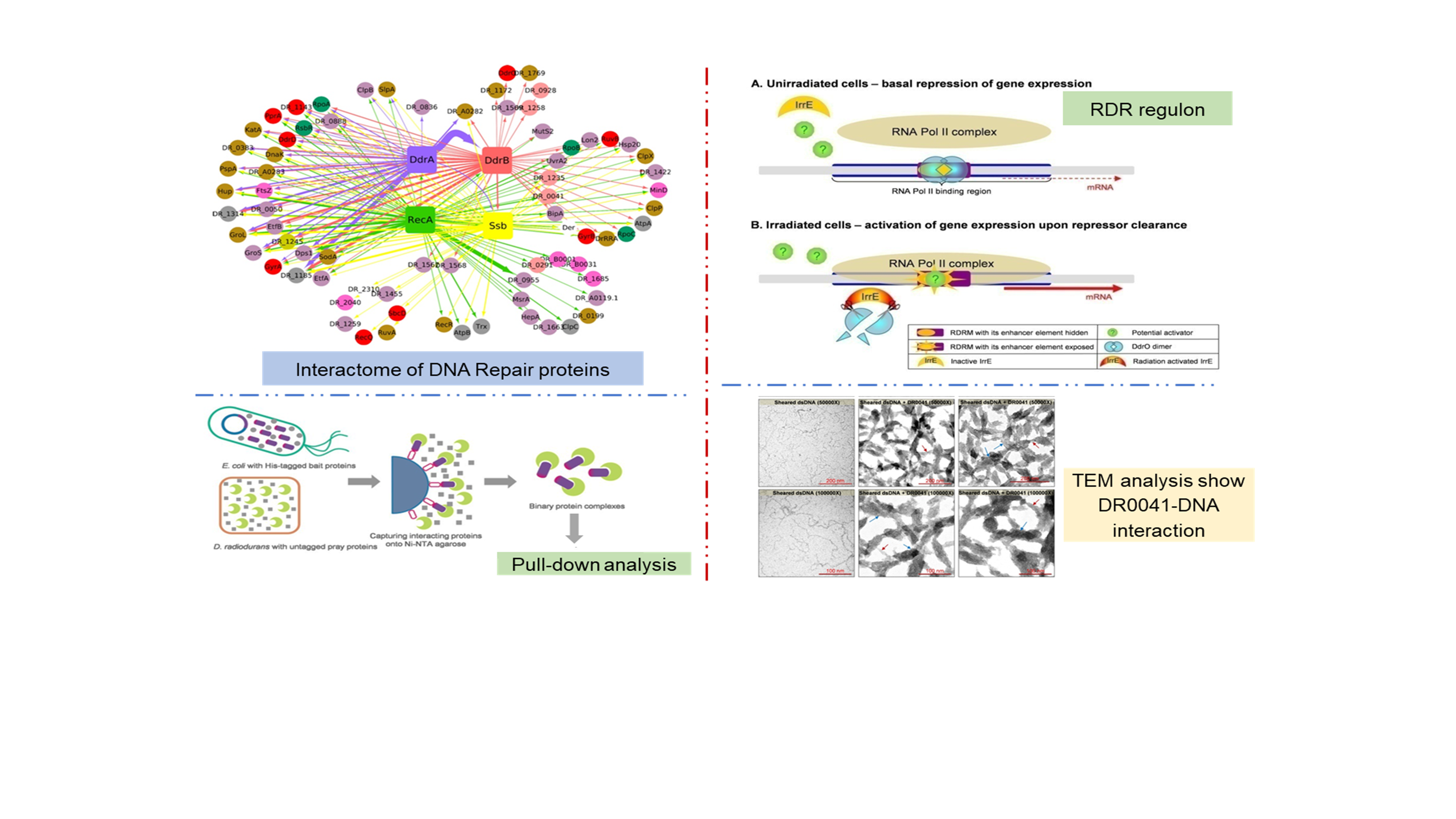
Understanding gene regulation, DNA-protein, and protein-protein interactions underlying DNA repair in <i>D. radiodurans</i>
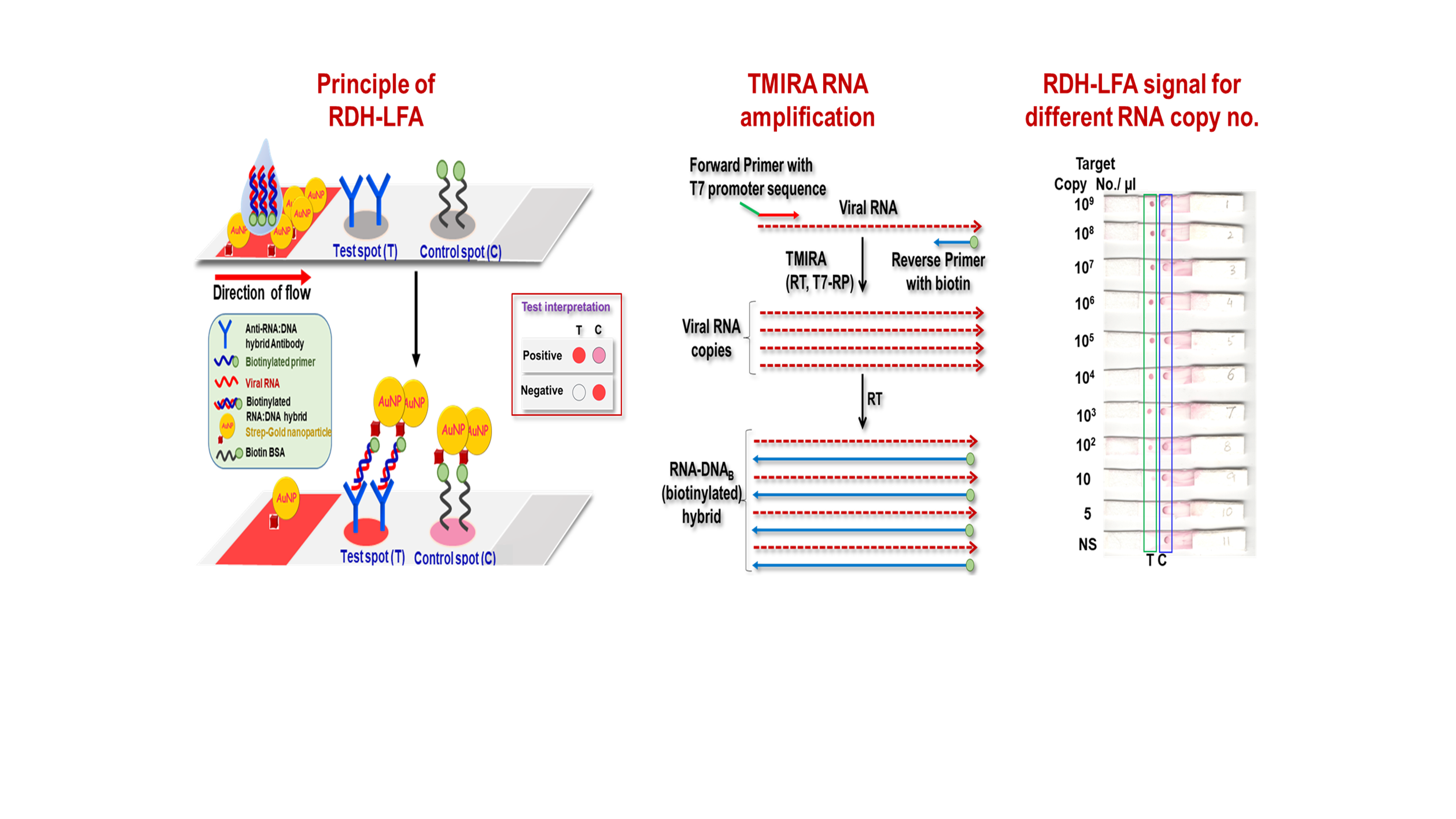
Development of a Rapid and Ultra-Sensitive RNA:DNA hybrid immunocapture based biosensor for visual detection of SARS-CoV-2